This is a running account of enhancements made to the Mastermind Genomic Search Engine. The most recent updates are at the top, followed by previous updates, so you can see all that the Genomenon team is doing to improve Mastermind in one place.
November 12, 2024
Our most recent changes to Mastermind include -
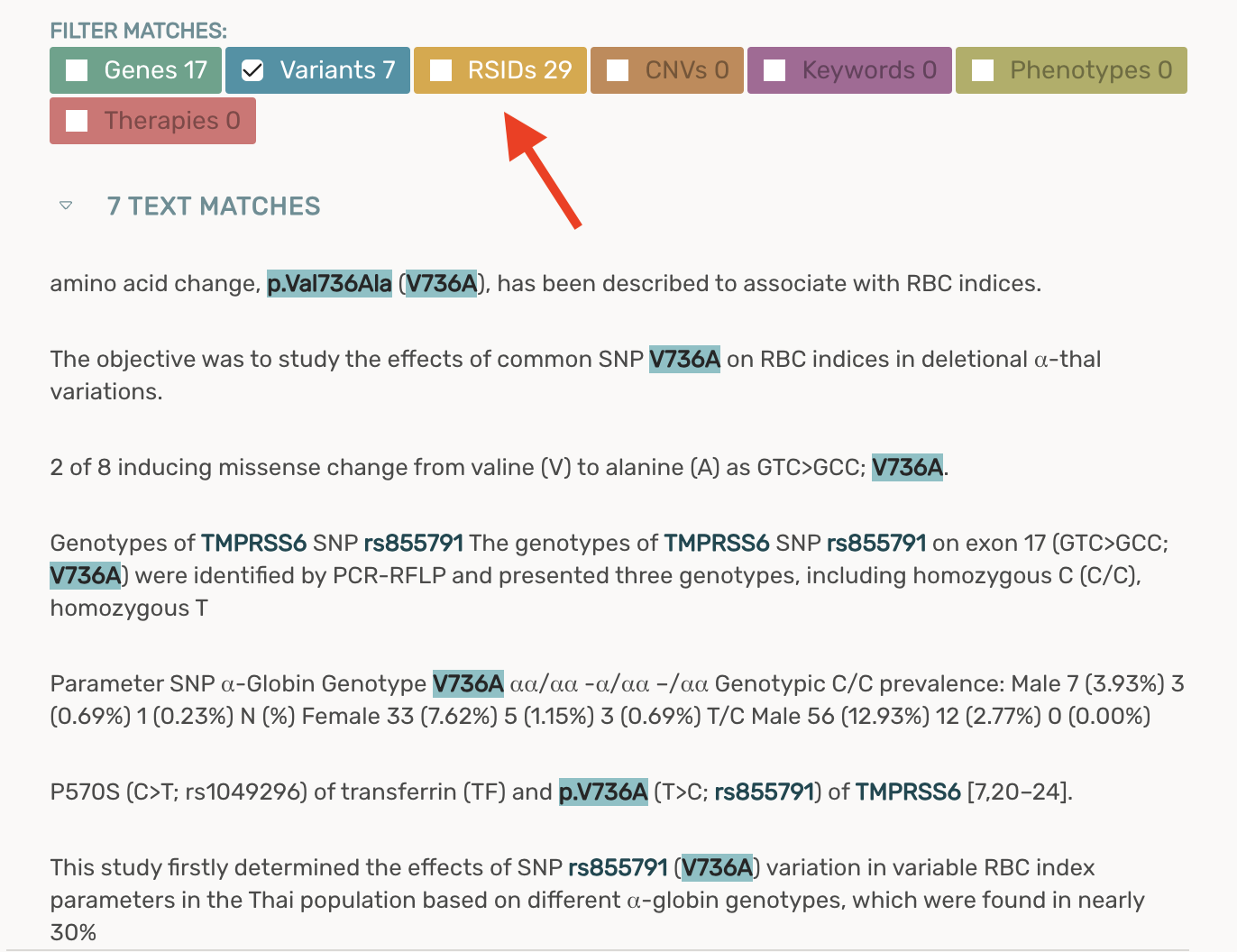
Adding a new rdID filter to the article viewer that enables you to add/remove rsID matches from your text fragments
- For maximum sensitivity, rsIDs are included in the text fragments by default
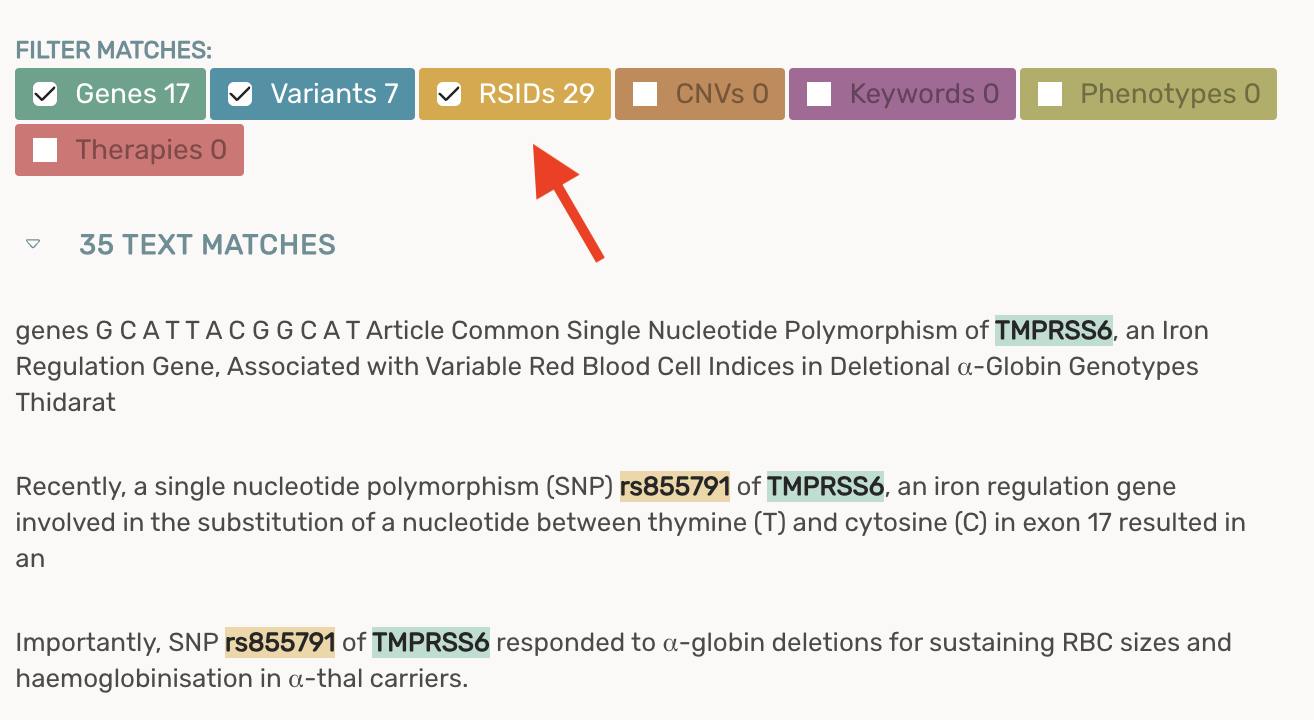
- Unchecking the rsID filter will remove rsID text fragments for all papers moving forward
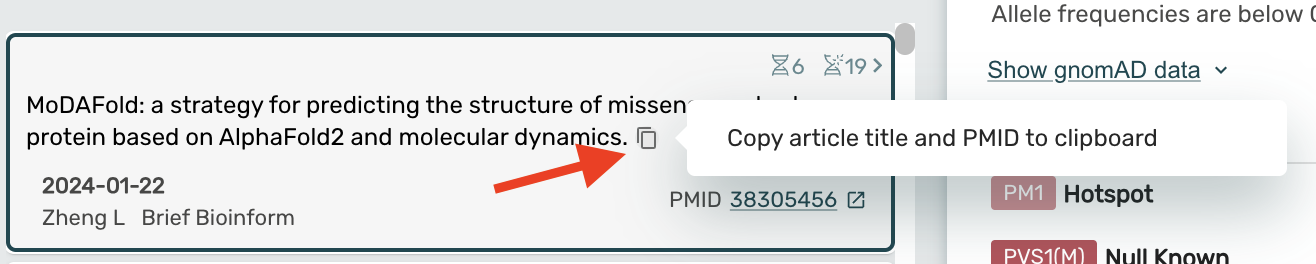
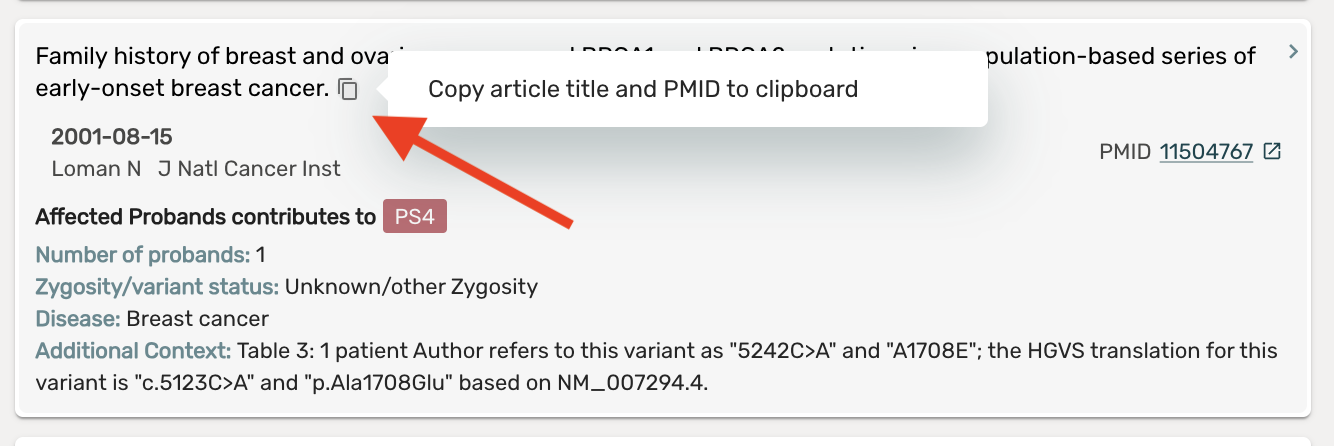
Enabled copying article title and PMID from any Article card to the clipboard
- Clicking the copy to clipboard button on any article card will automatically copy the title and PMID
Access to the GTEx portal from the gene page for all genes
- Use the GTEx link out in the Gene Summary Section of the Gene Page
Please feel free to reach out to support@genomenon.com if you have any questions about the recent changes to Mastermind, or if you have any feedback on features or changes that you would like to see in Mastermind!
October 23, 2024
The Genomenon team is excited to announce the new and updated Curated Evidence Viewer with this latest release of Mastermind!
In this release, we have -
- replaced the interpretation page with the newly designed Curated Evidence Viewer and seamlessly integrated with the Articles page
- updated our curation methodology to include additional data and increased transparency in how the data is displayed
With these enhancements, users will benefit from a range of valuable improvements, including:
- Improved transparency in the evidence identified in each article and for the variant.
- Enhanced readability of the user interface.
- Greater flexibility in the application of ACMG criteria based on the entirety of the data
- Increased ability to modify the criteria strength in light of new gene/disease specific guidelines are published.
- More detailed information on curated articles that contain no additional evidence for classification.
- Access to the most accurate and up-to-date evidence in their variant interpretations leveraging the REVEL protein prediction model.
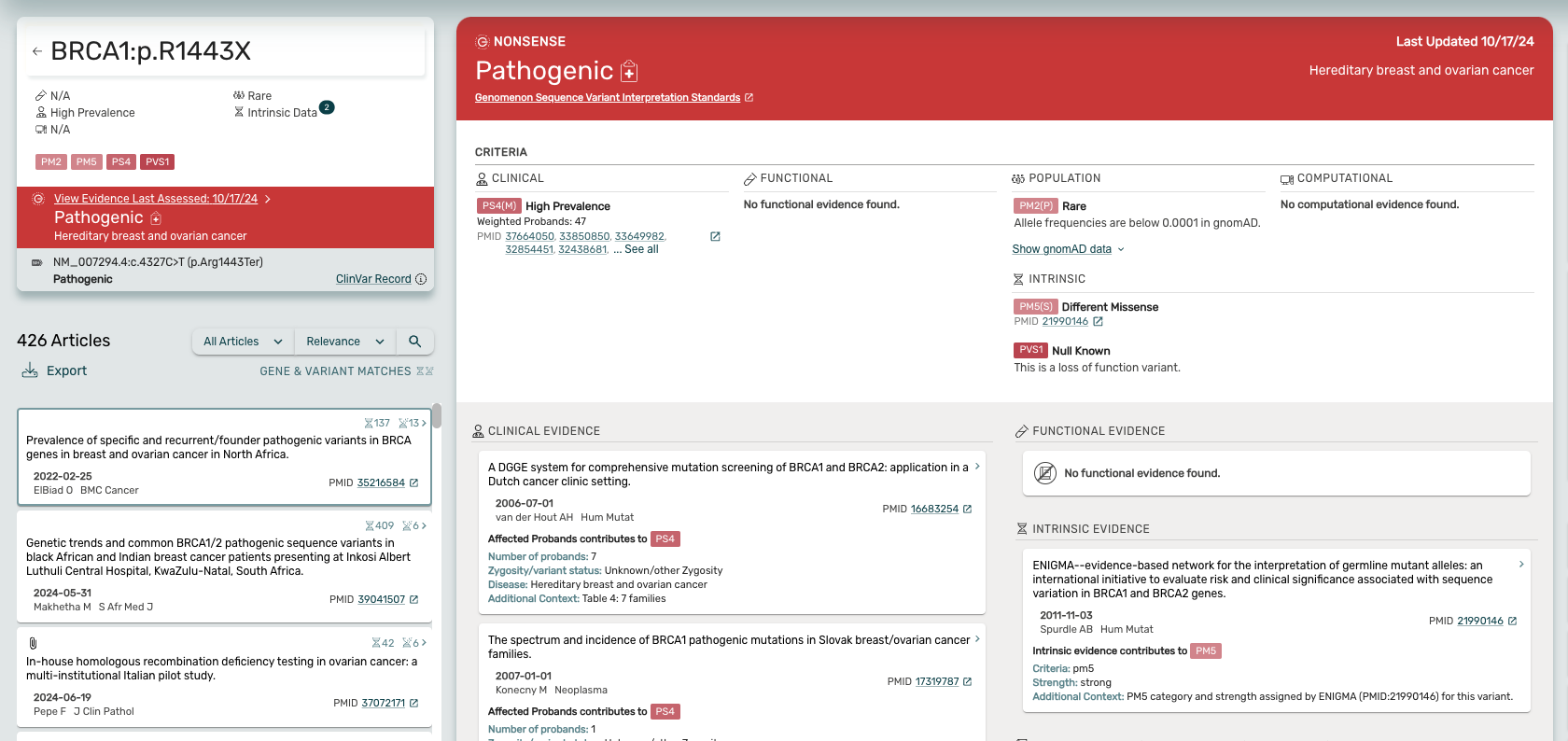
The Curated Evidence Viewer -
Access detailed insights with unmatched ease!
- Users can access the Curated Evidence Viewer without needing to click “View Interpretation” or launch a new page.
- When searching for a variant curated by the Genomenon team, users will land directly on the Curated Evidence viewer, showcasing all the articles and discrete data that contribute to the classification.
- Navigate between the curated evidence and articles by clicking on the colored classification bar in the variant card.
- Clicking an article card in the article list or the Curated Evidence Viewer enables users to see the text fragments for a given article.
- Return to the Curated Evidence Viewer by clicking the colored classification bar in the variant card located at the top left of the page.
Variant Information Card -
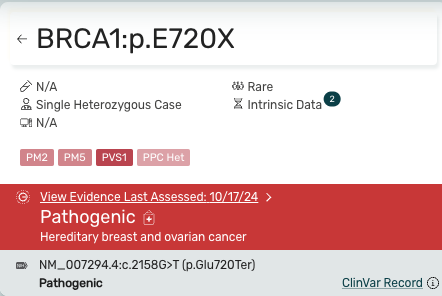
- The icons and data at the top provide a concise summary of the curated evidence.
- Users can toggle between text fragments and the Curated Evidence Viewer by alternatively clicking between the classification banner and the article card.
- The ClinVar record is shown below the pathogenic banner for additional context.
ACMG Criteria Summary Section -
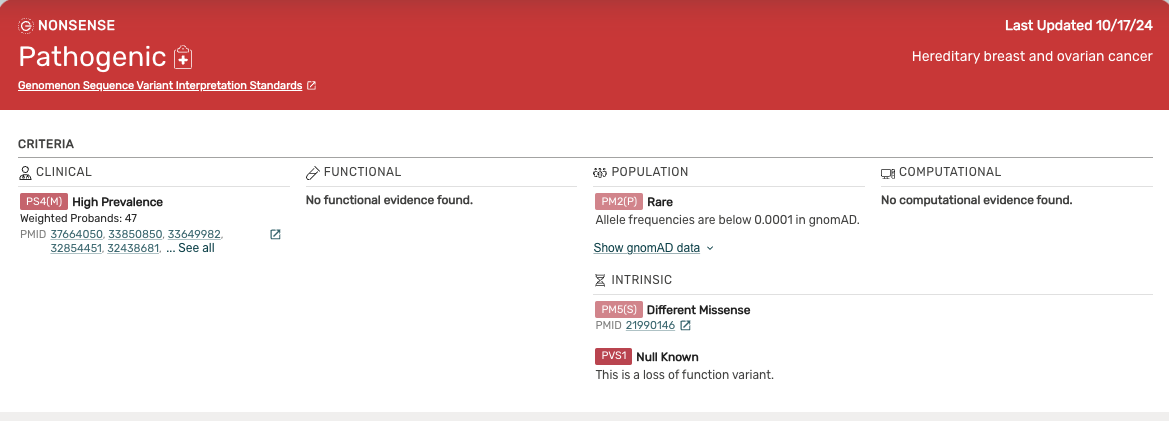
- The top part of the Curated Evidence viewer is the ACMG Criteria summary section, and shows all of the curated literature and information that contributes to an ACMG criteria.
- The summary section breaks evidence into Clinical, Functional, Population, Intrinsic, and Computational. This section only shows weighted evidence.
- Clicking on a PMID in the summary will open a new tab for that paper in PubMed.

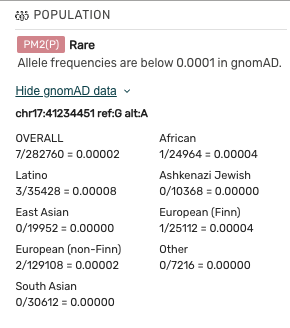
- Clicking the “Show gnomAD Data” drop will display subpopulation data. Mastermind currently uses gnomAD version 2.1.1.
Curated Evidence Article List and Article Cards -
- Below the ACMG Criteria Summary Section, ALL articles that were evaluated for this variant will be displayed.
- Articles and discrete data that directly contribute to an ACMG criterion are shown along with the corresponding ACMG criterion.
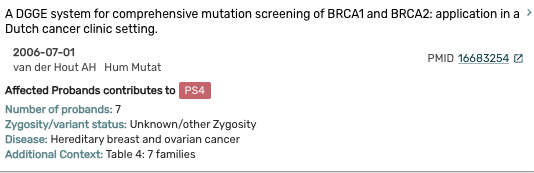
- Also included are articles that were evaluated but did not rise to the level of weighted evidence. These articles are displayed in the Variant Referenced With Inapplicable Evidence Section.

- Clicking a PMID on an article card within the Curated Evidence Article List will open the article in PubMed.
- Clicking elsewhere on the article card will bring the user back to the text fragments.
Curation methodology -
Enhanced evidence for evolving standards.
In response to user feedback and in anticipation of the changes to the ACMG sequence interpretation guidelines, we have made significant improvements to our curated evidence.
Key enhancements include -
- Providing discrete data curated by our expert team of variant scientists.
- Data is included for individual article curations and at the variant level.
- Integration of REVEL into our computational ACMG criteria. REVEL utilizes an ensemble method specifically designed to predict the pathogenicity of rare missense variants. By combining scores from a diverse set of predictive tools, REVEL achieves superior performance compared to other models, offering a more reliable and comprehensive assessment.
Please feel free to reach out to support@genomenon.com if you have any questions about the recent changes to Mastermind, or if you have any feedback on features or changes that you would like to see in Mastermind!
Best,
The Mastermind Team at Genomenon
June 12, 2024
The Genomenon team is excited to announce upcoming changes to the Mastermind Genomic Intelligence platform.
Please see below for the most recent updates:
- Our Genomenon curation experts recently added new Gene-Disease Relationships with Curated Evidence
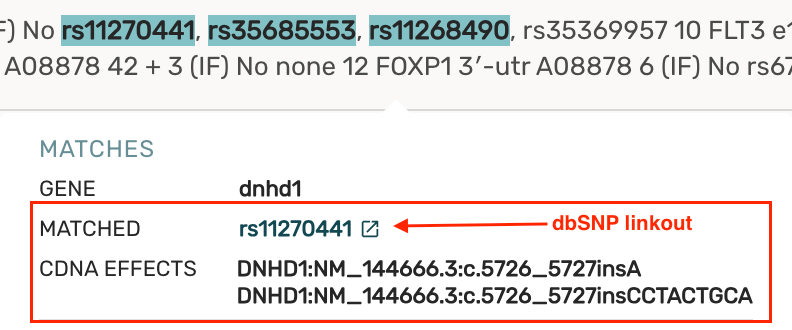
- For rsID text fragments, we include any cDNA effects and a linkout to dbSNP
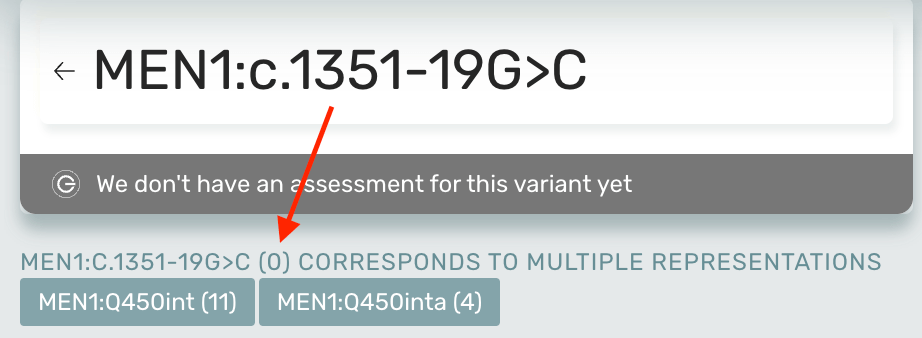
The Mastermind articles page now displays the number of article matches for the exact nomenclature searched and for multiple representations:
- For example, a search for MEN1:c.1351-19G>C is also represented by MEN1:Q450int and MEN1:Q450inta.
- We display the number of article matches for MEN1:c.1351-19G>C (0 articles) and MEN1:Q450int (11 articles), and MEN1:Q450inta (4 articles)
- For more information about multiple representations for non-coding variants, check out our FAQs
In response to user suggestions, we have implemented several enhancements that improve the article viewing experience:
- Two-pane scrolling enables maximum viewing area for articles and text fragments
- All text fragments are displayed upon opening an article
- Filter text match selections persist as you navigate through articles

- Users with Mastermind Basic edition will see reduced functionality, including being unable to view calculated gene information and articles sorted by relevancy score.
- Users with Mastermind Basic Edition can request a free 7-day trial to the Professional edition by clicking on the “Request a 7 day trial” button on the navigation bar in the Mastermind platform.
Please feel free to reach out to support@genomenon.com if you have questions about this release, or feedback on features or changes that you would like to see in Mastermind!
April 9, 2024
Please see below for the most recent changes to Mastermind 3.1:
NEW Mastermind Homepage and Search Bar
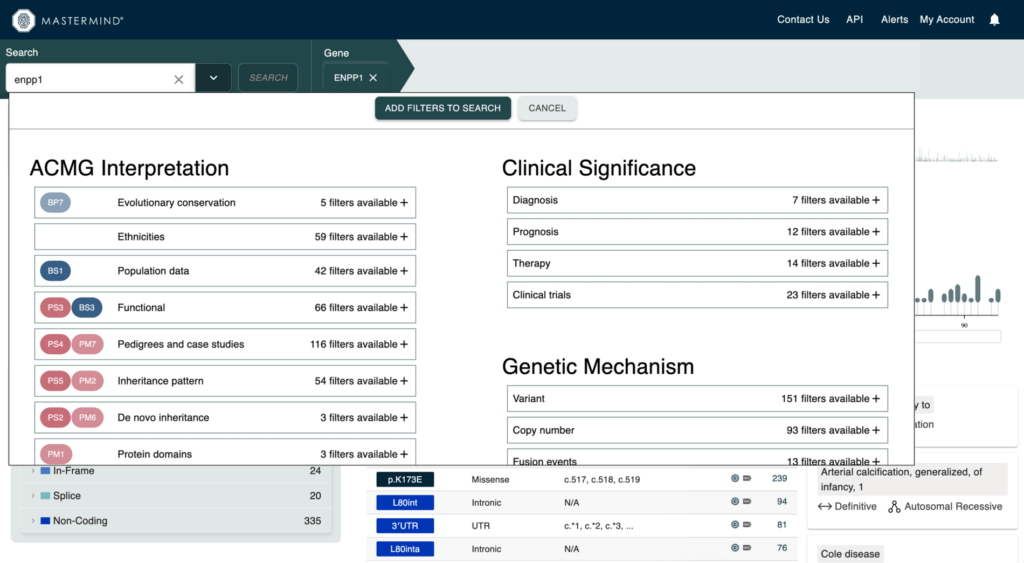
Adding keywords to your search can be done from the homepage now!
- Click the caret on the search bar to pull up the filter keyword interface
- Select a keyword to filter your article list
- Click “Add Filters to Search” to apply the keyword to your search and the selected Keywords will be present in the search bar

Users can still take advantage of the Multi-Entity Search Parsing with new + button in the new search bar.
- Enter a string of text with Genes, Variants, Diseases, Phenotypes, and Therapies and Mastermind will recognize all terms and display suggestions below the search bar. By holding down shift, OR clicking the NEW + button you can select and add every relevant search term to the search bar.
- Other features like boolean searching and tag-editing are still available for users on the new search bar

NEW Evidence Page
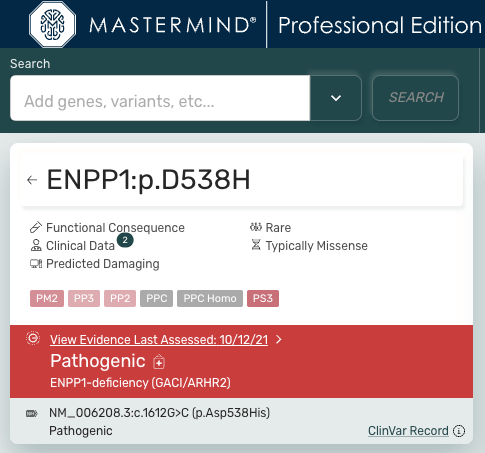
The Evidence Page is a redesigned version of the Focus Page with all the classic Mastermind features, and an abundance of new features. When searching a variant, the Variant Information Card will be present in the top left of the page
- Genomenon curated evidence and/or a ClinVar record will be show in the top left of the page on the variant info card
- For Genomenon Curated Evidence Preview, we now display the ACMG categories used to curate the variant
- Click the “View Evidence Last Assessed” to see the curated evidence page
- The ClinVar record for a variant is below the Genomenon Curated Evidence section. Click the ClinVar Record hyperlink to link out to ClinVar, hover over the ‘i’ icon to see a preview of the ClinVar Record. Next, click the arrow next to the Gene to navigate to the Gene Page
When viewing evidence for a gene, the Gene Info Card will be present in the top left of the page
- GDRs will be displayed below the Gene name while viewing evidence
- Click the arrow next to the Gene to navigate to the Gene Page
The Article List is now on the left side of the page
- We are thrilled to bring two highly requested features to the article list
- A Genomenon Symbol with “Clinical Evidence” or “Functional Evidence” tag will be present on articles in the article list that were used for curate the variant of interest
- Articles that are present in the ClinVar record will display an NIH symbol next to the PMID

Text Matches, Supplemental Matches, and the Abstract are now front and center of the page
- Text matches will now be color-coded
- Click the color-coded text match in order to see the matched nomenclature

Related Variants is now on the right hand side of the page and Mastermind now displays pathogenicity for related variants if curated.
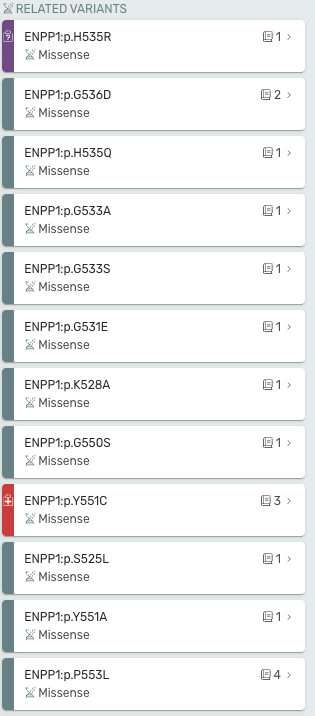
Gene Page Updates
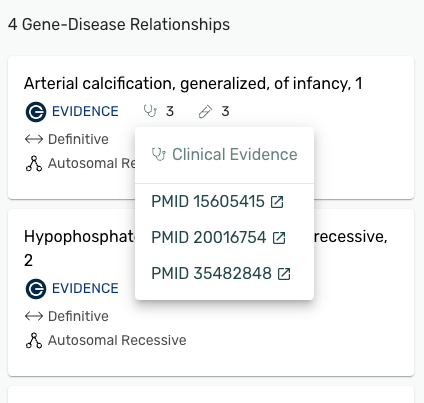
Evidence for Gene-Disease Relationships (GDRs) is now available on the gene page! Mastermind provides a curated list of clinical evidence (represented by the stethoscope icon) and functional evidence (represented by the test tube icon) our curation team used to establish the Gene-Disease Relationships
- Clicking on the number next to the symbol will pop up the list PMIDs
- Clicking the PMID will open the article in PubMed
- Use the filter drop down in the variant table to see which variants our curation team at Genomenon curated, and also see which variants have a ClinVar Record.
We have helpful video tutorials about these updates that you can view here.
Please feel free to reach out to support@genomenon.com if you have any questions about the recent changes to Mastermind, or if you have any feedback on features or changes that you would like to see in Mastermind!
Still need a Mastermind account? Sign up here and get started with a complimentary trial of Professional Edition!
November 2, 2023
Mastermind 3.0 Release: Gene Information Page
Over the last year, the dedicated team at Genomenon have been working tirelessly to understand what users want and need from Mastermind. Today, we are launching the first step toward an enhanced user experience and user interface with the release of Mastermind’s new Gene Information Page.

We are thrilled to give users a taste of the new interface that will continue to change over the next year. As we continue to work to bring you the best version of Mastermind, you will notice a combination of new and old pages. Your feedback will help drive the development of current and future enhancements coming to Mastermind.
What is the Gene Information Page and how do I get there?
- The Gene Information Page is a central location with crucial information for understanding the gene from various sources such as ClinVar, ClinGen, DECIPHER, and gnomAD
- Any single gene search combined with a disease, phenotype, therapy or category keywords brings you to the gene page
What info is available on the Gene Information Page?
- Curated Gene-Disease Relationships (GDRs)
- Our curation team develops GDRs based on ClinGen guidelines and presents the disease, strength of association, and inheritance pattern for definitive GDRs
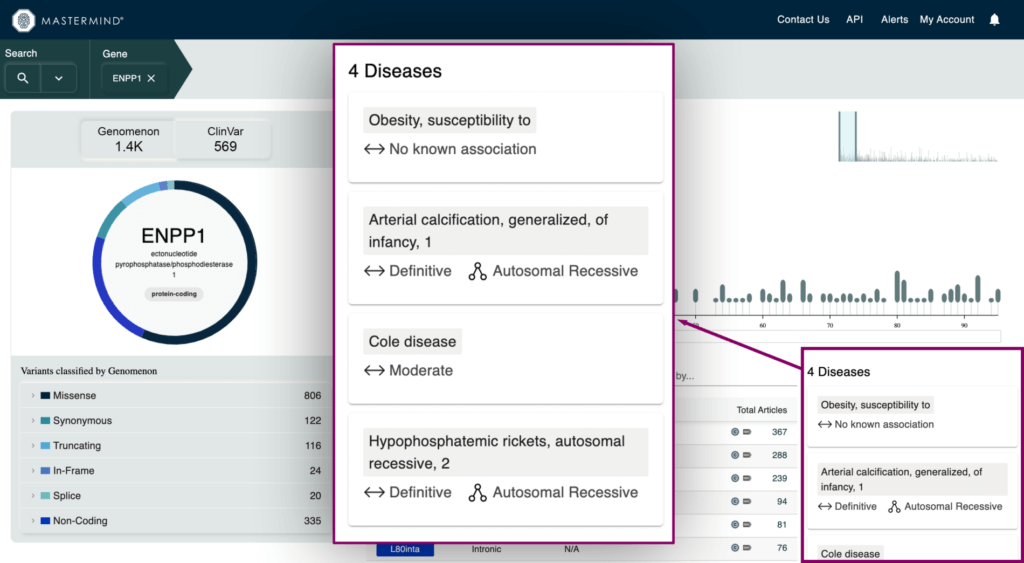
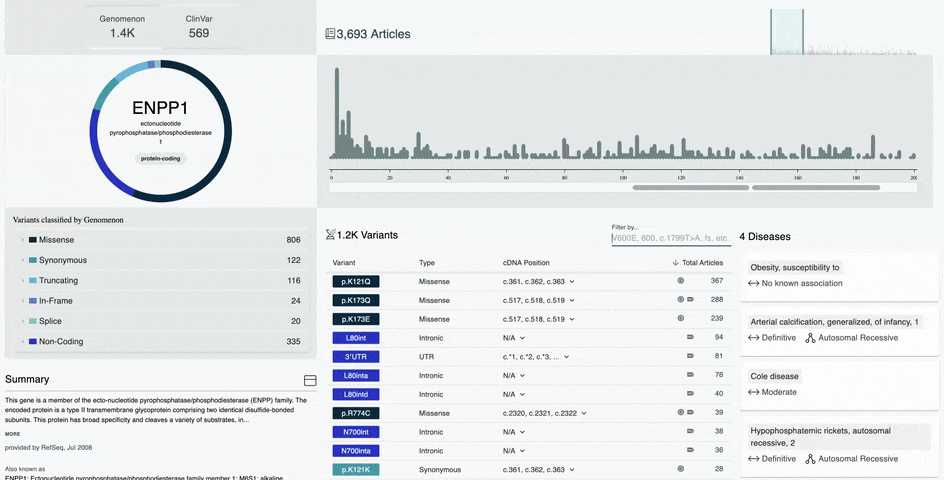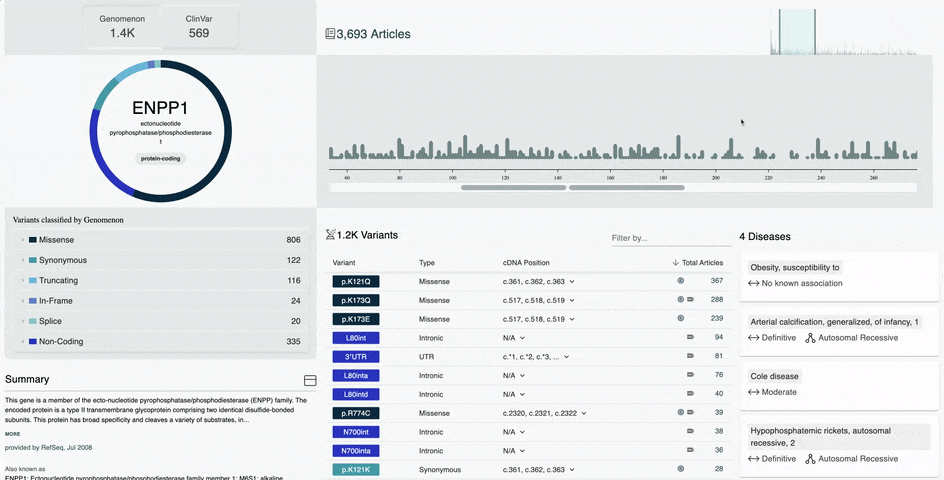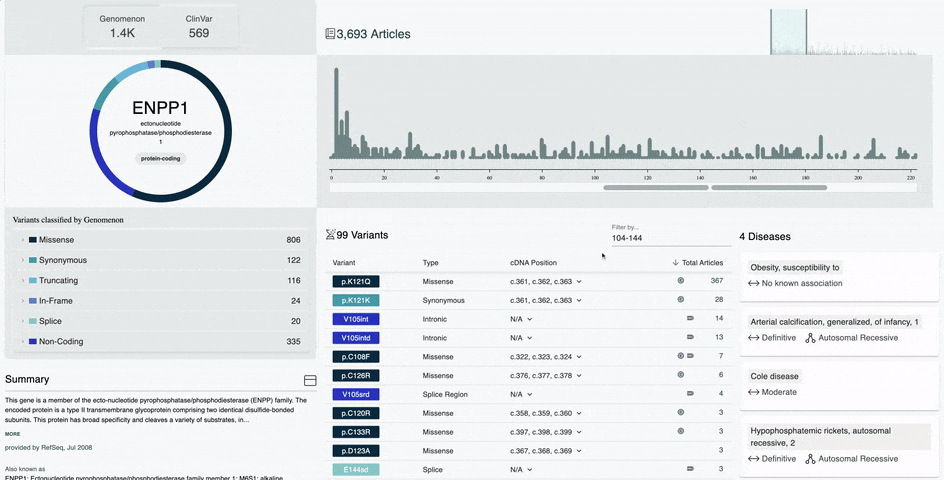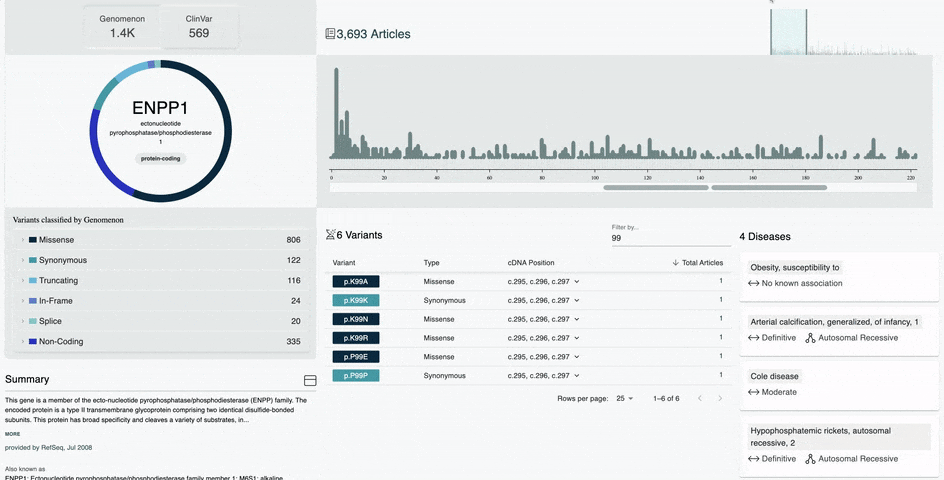
- Variant Type and Pathogenicity Breakdown visual for Genomenon and ClinVar variants
- Metrics for total variants in a gene broken down by variant type and pathogenicity
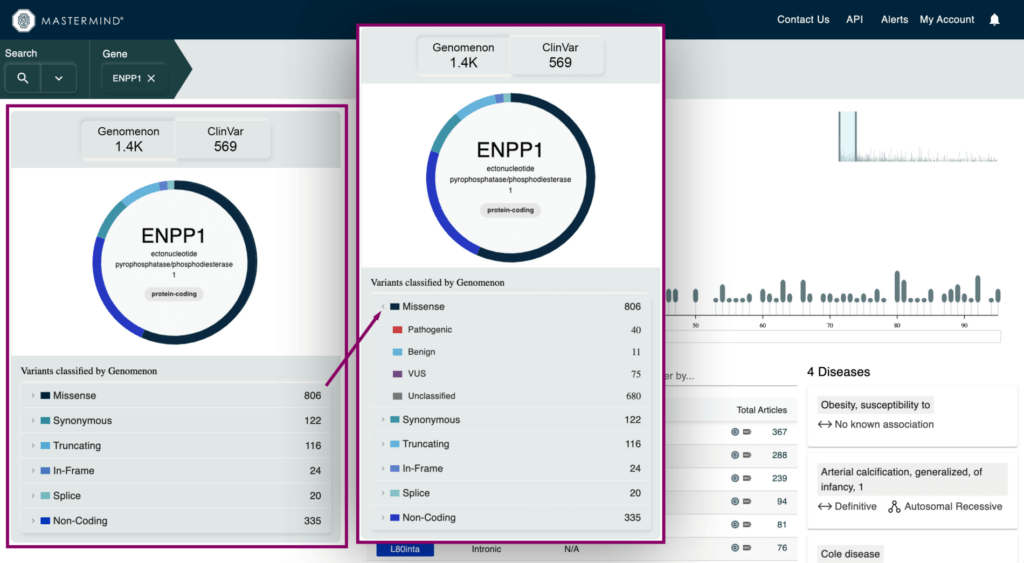
- Gene Summary information
- High level gene information such as description of the gene, canonical transcript, chromosome location, protein shifts, and critical protein domains
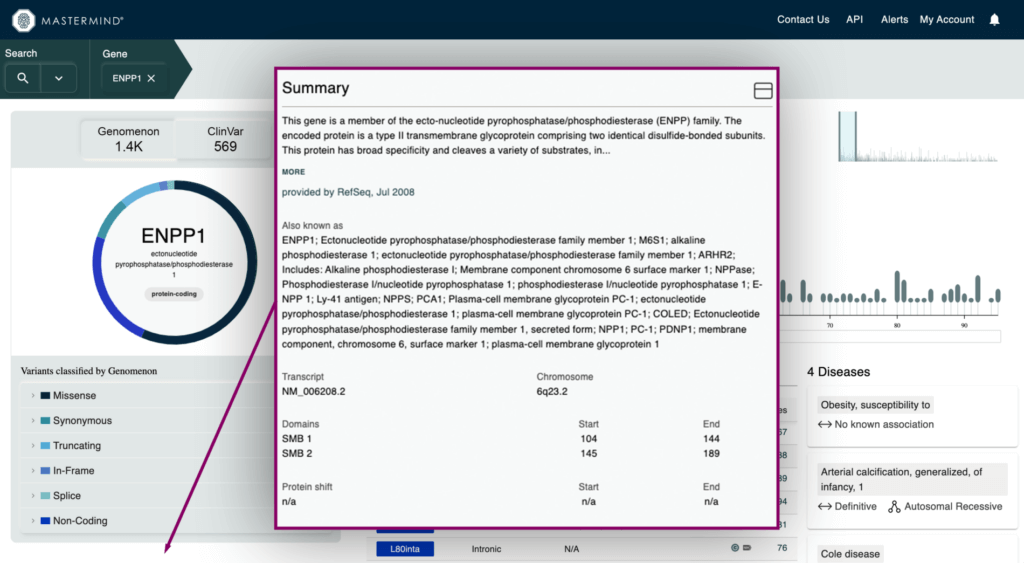
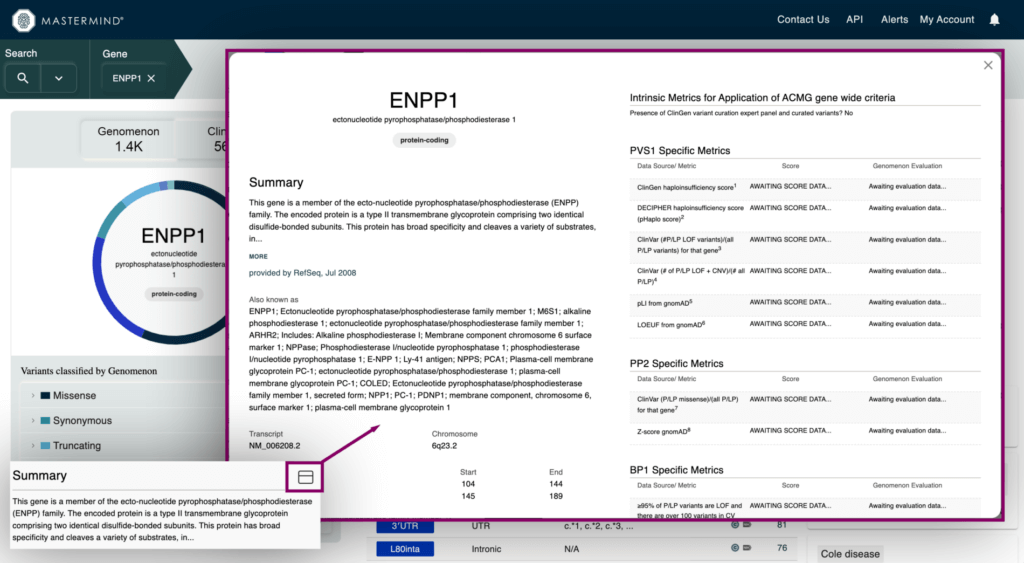
- Intrinsic Metrics for Application of ACMG Gene-Wide Criteria
- Information from ClinGen, ClinVar, DECIPHER, and gnomAD that applies to curation based ACMG guidelines
- Haploinsufficiency score from ClinGen and DECIPHER, pLI and LEOUF from gnomAD, and much more
What enhancements were made to current Mastermind features?
- Improved Variant Diagram on the Gene Information Page
- Hover over the amino acid position in the diagram to view the number of variants and most researched variants at that position
- Simply click that amino acid position to view all variants at that position in the table below
- Hover over a protein domain on the x-axis to view the domain name and amino acid position range
- Simply click on that protein domain to view all variants in that domain in the table below
- Improved Variant Table on the Gene Information Page
- Filter by any amino acid position range in the variant table
- Sort by variant position, type, cDNA position, and total articles
- Variants curated by our team will display a Genomenon icon in the table
- Variants with a ClinVar record will display an NIH icon in the table
How do I search the new Gene Information Page?
- View articles for a variant by entering the variant in the search bar or clicking on the article count in the variant table
- The filter categories are located next to the search magnifying glass and represented by the dropdown arrow
- Any search with multiple genes or a CNV will bring you to the classic view of Mastermind
- Any search containing a variant will bring you directly to the articles for that variant
Who has access to the Gene Information Page?
- All users have access to the the Gene Information Page
- Mastermind Basic users have access to some information for all genes and all information for a limited number of genes (try a test gene)
- Mastermind Pro users have unlimited access to all the Genomenon curated evidence, pathogenicity breakdown, and current Pro features like category keywords on the Gene Page
Please feel free to reach out to support@genomenon.com if you have any questions about the recent changes to Mastermind, or if you have any feedback on features or changes that you would like to see in Mastermind!
July 11, 2023
Mastermind Feature Release: Multi-Entity Search Parsing
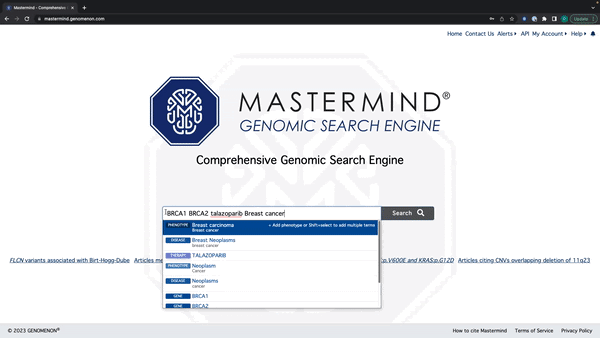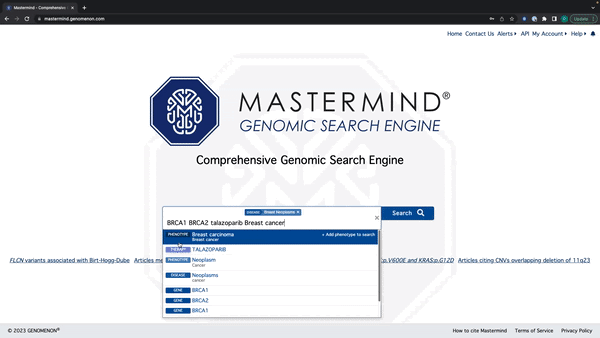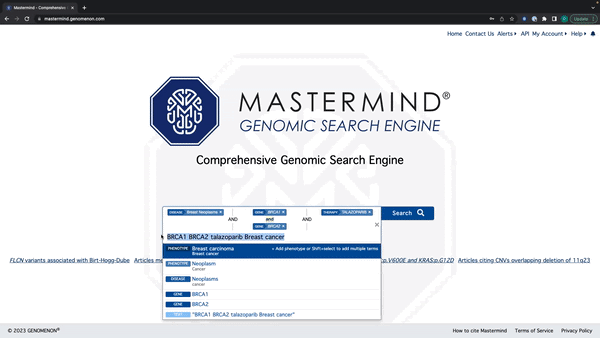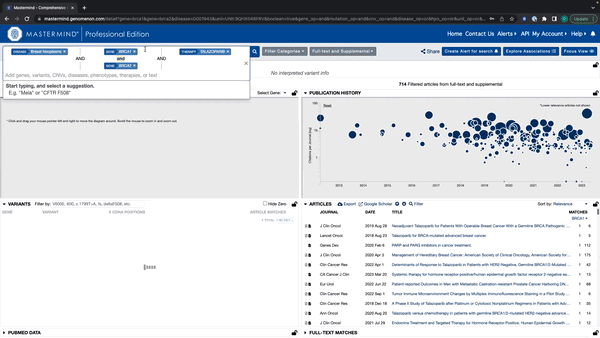
- Mastermind will now recognize all search terms in a string of text
- Enter a string of text with Genes, Variants, Diseases, Phenotypes, and Therapies
- Mastermind will recognize all terms and display suggestions below the search bar By holding down shift, you can select and add every relevant search term to the search bar
Watch this quick tutorial video of how to use this new feature.
Mastermind Feature Release: Tag Editing
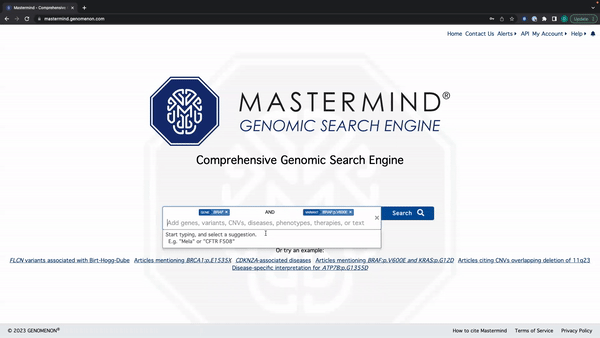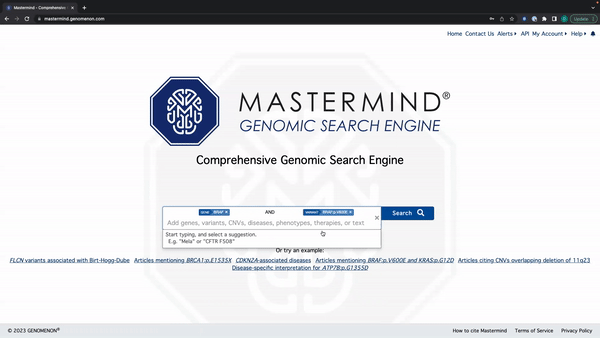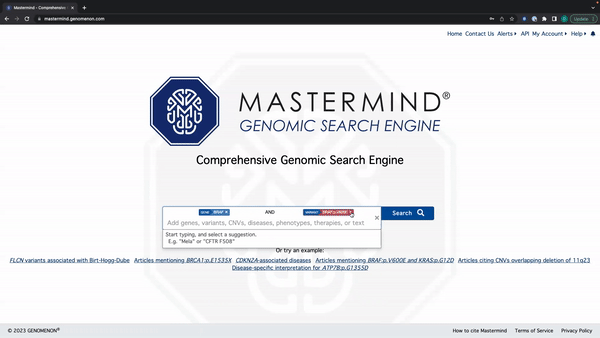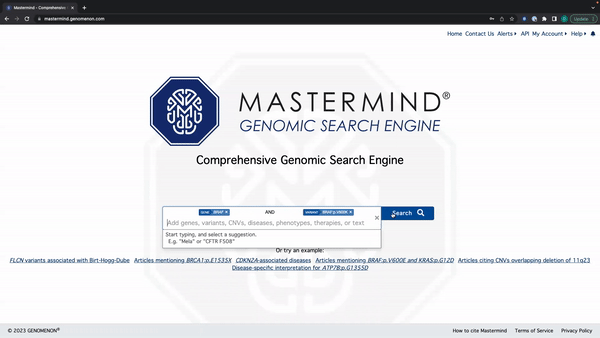
- You can edit the search tag by clicking anywhere except for the X on the tag
See the video here for a tutorial using the example: BRAF and BRAF:p.V600E.
Please feel free to reach out to support@genomenon.com if you have any questions about the recent changes to Mastermind, or if you have any feedback on features or changes that you would like to see in Mastermind!
May 16, 2023
Mastermind Feature Release:
Supplemental File Enhancements
The Mastermind team is excited to announce that we have made several enhancements to our supplemental file matching to help you save time and augment your genomic evidence search experience. This blog highlights these new features in more detail, and I encourage you to watch the quick tutorial video below to see how it works.
We know that one of the features users value most is being able to see whether a variant is cited in supplemental materials, something Mastermind excels at. We also know that finding the variants in the supplemental files can be cumbersome and time-consuming. Our customer success and product development teams have been working tirelessly to provide a solution for this critical pain point.
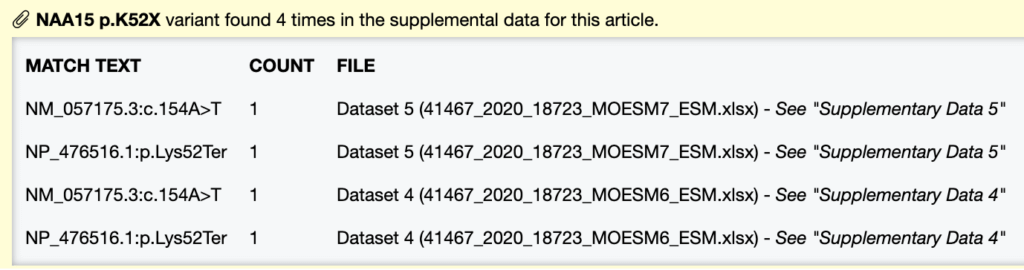
Historically, Mastermind only showed that your variant was found in the supplemental file. Moving forward, Mastermind will now display the exact variant nomenclature found in the supplemental file, along with the number of times that specific nomenclature was mentioned. In addition, we will provide the exact supplemental file name and tab to allow you to easily navigate to the supplemental file match.
For example, if you perform a search for ATP7B:p.G1355D and Mastermind recognizes c.4064G>A twice in the supplemental file, we will display that the cDNA nomenclature (c.4064G>A) for ATP7B:p.G1355D was found 2 times in Supplementary File 1, in the excel tab “Table S2”.

By providing this information, you are able to copy your nomenclature match, navigate to the supplemental file and tab, and use the search function to easily find your variant.
Release Notes Summary
Mastermind now provides the following additional features to our supplemental matches for users with Mastermind Professional Edition:
- Display the exact nomenclature matched in the supplemental files along with the number of matches
- Display the supplemental file name where each match was found
- Display the exact tab of the excel file where the match was found
See example below:
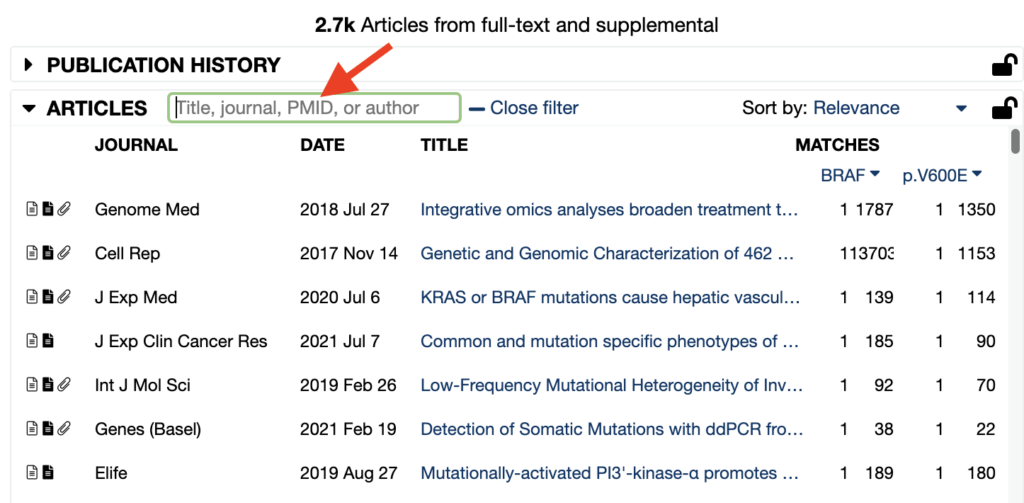
Another new feature: Filter by PMID
Looking for a specific paper? In the Articles pane, users are now able to search for a specific PMID in the article list to find what you’re specifically looking for. The ability to filter by PMID in the article list is available for all Mastermind Users.
Take a quick tour of the updates:
With the latest update, the Mastermind Genomic Search Engine is poised to deliver even more value for our users by empowering them to identify the most relevant genomic data for their workflows. Log in or create your free Mastermind account today to see these latest features for yourself.
Get in touch with us.
Please feel free to reach out to support@genomenon.com if you have any questions about the recent changes to Mastermind, or if you have any feedback on features or changes that you would like to see in Mastermind!







